
INTRODUCTION
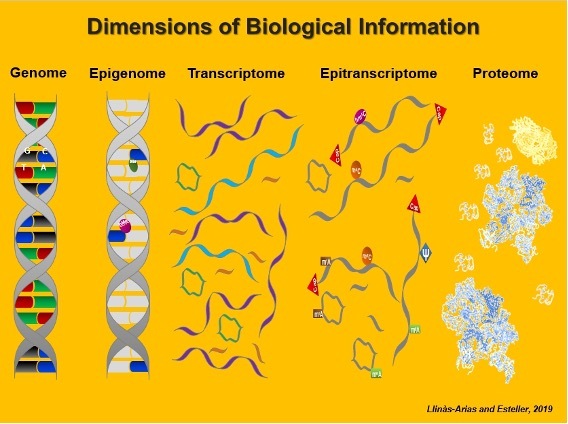
OUR RESEARCH
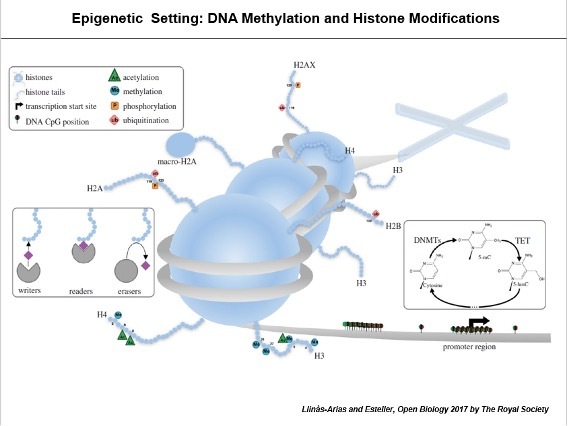
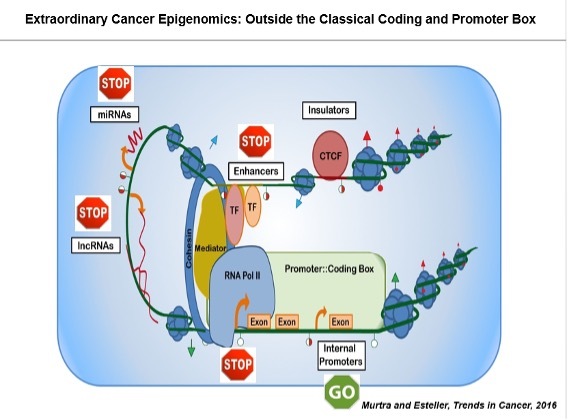
Our laboratory is one of those responsible for establishing the observation that epigenetic disruption of mRNA transcription, particularly in DNA methylation and histone modification patterns, contribute to the initiation and progression of human tumours (reviewed in Esteller, N Engl J Med 2008; Heyn and Esteller, Nat Rev Genet 2012; Berdasco and Esteller, Nat Rev Genet 2019; Davalos and Esteller, CA Cancer J Clin 2023).
It has also been recognized that microRNAs (small non-coding RNAs that regulate gene expression by sequence-specific base pairing in mRNA targets) also play a key role in the biology of the cell, and can have an impact on the development of cancer. In this context, we characterized the first miRNA undergoing specific cancer-methylation associated silencing (Lujambio et al., Cancer Res 2007), followed by the characterization of many other miRNAs disrupted in the same manner (Lujambio et al., PNAS 2008; Davalos et al., Oncogene 2012).
OUR GOALS
Our group has had a long-standing interest in translating the use of epigenetic knowledge gained from research into biomarkers to predict clinical outcome and to assay new drugs to reverse the distorted epigenetic landscape (Berdasco and Esteller, Nature Review Genetics 2019). For example, we have used epigenetic markers to predict response to anti-tumour therapies and following the initial observation that MGMT gene methylation predicted response to alkylating agents in glioma (Esteller et al., N Engl J Med 2000).
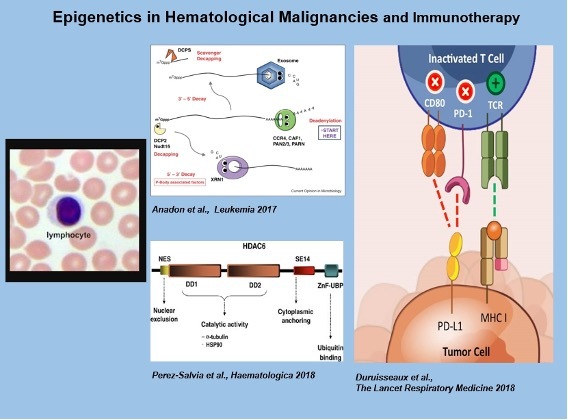
OUR CHALLENGES
Continuing with this translational side of our work, we are also interested in the development and study of new epigenetic drugs that target DNA methylation and histone modification writers, readers and erasers and could have an anti-cancer effect (Lara et al Oncogene 2008; Zubia et al Oncogene 2009; Huertas et al., Oncogene 2012; Perez-Salvia et al., Oncotarget 2017; Perez-Salvia et al., Haematologica 2018).
Team

Senior group leader

Research Associate

Research Associate

Research Associate

Research Associate

Postdoctoral Investigator
Postdoctoral Investigator

Postdoctoral Investigator

Postdoctoral Investigator

Postdoctoral Investigator

Postdoctoral Investigator

Postdoctoral Investigator

PhD Student

PhD Student
PhD Student

PhD Student

PhD Student

PhD Student

PhD Student

PhD Student

Senior Lab Technician
Selected Publications
Current Grants
Ministerio de ciencia e innovación
COMBATIR NUEVAS ESTRATEGIAS COMBINATORIAS Y BIOMARCADORES DE RESPUESTA A INHIBIDORES DE HDAC6 PARA EL TRATAMIENTO DEL CÁNCER.
European academy of dermatology and venereology
SCIENCE-SS Single-cell Characterization of Epigenetic and Transcriptomic Dynamics in Sézary Syndrome Progression
Fundació "la caixa"
Solve-ALD Brain Inflammation in adrenoleukodystrophy: from drivers to treatments
Instituto de salud carlos iii
SEED-ALS Synergizing Efforts to Develop and Accelerate Breakthroughs in ALS Research
Gilead sciences s.l.
Identification of DLBCL key molecular features for CAR-T cell therapy
Fundació "la caixa"
Solve-ALD Brain Inflammation in adrenoleukodystrophy: from drivers to treatments

Fundación científica de la asociación española contra el cáncer
PREDICT-Meso PRE-malignant Drivers Combined with Target-Drug validation in Mesothelioma

European commission
SENESCENCE2030 Targeting Cell Senescence to Prevent Age-Related Diseases
Deutsche josé carreras leukämie stiftung
scEpiCART Refinement of Epigenetic Factors to Improve CAR T Cell Therapeutic Efficacy
Fundación mutua madrileña
Identificación de determinantes de respuesta a quimioinmunoterapia en biopsia líquida, en pacientes con cáncer microcítico de pulmón.

Ministerio de ciencia e innovación
ASPIRE Neoadjuvant-adjuvant immunotherapy and cancer vaccines to improve survival in hepatocellular carcinoma
Instituto de salud carlos iii
CIBERONC Centro de Investigación Biomédica en Red Cáncer
Cll global research foundation
Gene Expression Regulation Study Mediated by the RNA-Binding Protein Musashi2 and the RNA Methyladenosine Transferase 3, as Therapeutic Targets in Chronic Lymphocytic Leukemia
Fundació "la caixa"
MyoClonal Somatic mutations and clonal hematopoiesis as predictors and drivers of heart failure progression

European commission
THRIVE TUMOUR-HOST INTERACTIONS IN LIVER CANCER OF CHILDHOOD AND ADULTS

Ministerio de ciencia e innovación
Single cell analysis of clonal heterogeneity in myelodysplastic syndromes treated with azacitidine

Ministerio de universidades
Estudios de epigenética del cáncer a nivel de célula única

Fundación científica de la asociación española contra el cáncer
T-REX Transcriptional Role and Epigenetics of the X chromosome in leukemias
European commission
IRB-TARGET IRB Barcelona International PhD programme: on TARGET for high-impact biomedicine

Agència de gestió d'ajuts universitaris i de recerca
Overcoming Cancer Immunotherapy Resistance

Ministerio de ciencia e innovación
USO DE APROXIMACIONES DE CELULA UNICA PARA DESCIFRAR LA EPIGENOMICA DEL CANCER Y LAS EPIDROGAS
