
OVERVIEW
In the interface between genomics, digital pathology, and artificial intelligence the Cellular Systems Genomicsgroup aims to define the spatiotemporal organization of complex tissues in health and disease, by the identification of key regulatory mechanisms driving heterogeneity in cellular identity and function, particularly in the context of inflammation, inflammatory disorders, and autoimmune diseases.
To address these questions, we will adopt a single-cell perspective, enabling the fine-grained and spatially resolved molecular profiling of tissues. We will develop new machine learning approaches and open-source tools to unlock molecular mechanisms hidden in large-scale datasets.
In a short-term perspective, these methods will help understand disease mechanisms, allowing the stratification of patients based on their molecular and cellular characteristics, ultimately providing new therapeutic targets for their treatments.
OUR RESEARCH
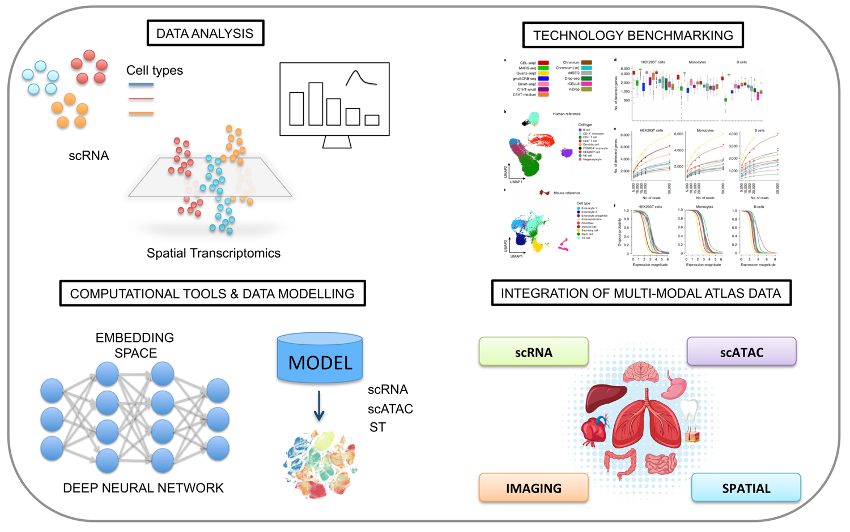
Single cell sequencing allows to profile thousands of individual cells per experiment, enabling the unbiased analysis of tissues, organs, and even entire organisms at an unprecedented resolution. These data represent a powerful tool for cell biology, with relevant clinical applications including diagnosis and treatment of diseases. Despite the many advantages of this approach, data are noisy and sparse, making the computational analysis challenging. To address these challenges, we apply machine learning and other statistical methods to develop new analytical frameworks and open-source tools to analyze, interpret and integrate data coming from single-cell and spatial genomics experiments.
As part of the Human Cell Atlas (HCA) consortium, which aims to create a catalogue of all cell types in our body, we have extensive experience on the systematic comparison of protocols in single cell RNA sequencing (scRNA-seq).
Beyond transcriptomic profiling with scRNA-seq, different cellular modalities can now be measured, including single-cell epigenetics (scATAC-seq), spatial transcriptomics as well as the joint profiling of chromatin accessibility and transcription on the same cell.
However, the integration of multimodal data poses new analytical challenges and new benchmarking are needed to assess reproducibility and integrity of these methods. We are working on new mathematical frameworks for the integration of multimodal data, enabling the comprehensive characterization of cells in their identity and function.

OUR GOALS
In the European Pancreas Atlas consortium (ESPACE, https://www.espace-h2020.eu), we are working to build a first version of the Human Cell Atlas of the Pancreas, by profiling the transcriptome and epigenome of cells from distinct anatomical regions of the adult pancreas. The integration of distinct single-cell and spatial data types will allow the comprehensive transcriptional and epigenetic landscape of pancreas cell types within their spatial context.
Our experience in single-cell data analysis on healthy and diseased tissues allowed us to build a deep understanding of cell-type structure and plasticity in different research contexts. To accelerate biological discovery and advance science, our group will share user-friendly computational solutions, by promoting open science, diversity and supporting an inclusive and collaborative environment. We welcome proposals for interdisciplinary research collaborations, from both industry and academia.
Selected Publications
Current Grants
Fundació internacional josep carreras
Decoding immune stages for enhanced CAR T-cell therapy in multiple myeloma
PID2022-137741OA-I00
Ministerio de ciencia e innovación
IMMUNOMYELOMICS Unveiling the single-cell transcriptomic and epigenomic regulation of T cell dysfunction in multiple myeloma treatment response (IMMUNOMYELOMICS).

RYC2021-032359-I
Ministerio de ciencia e innovación
Targeting inflammation in the era of single-cell genomics




